RNA alternative splicing is a crucial biological process that generates multiple variants of a single gene from a common primary transcript. This mechanism allows for the production of numerous protein products by selectively including or excluding specific exons during the formation of the final mature RNA. The regulation of alternative splicing involves a highly coordinated interaction between trans-acting factors, such as splicing regulators, and cis-acting elements, including splice sites, exonic and intronic enhancers, and silencers. These elements work together to determine whether particular exons are included or excluded in the resulting mRNA, shaping the ultimate protein-coding potential of a gene.
Alternative splicing is widespread in eukaryotic organisms and is estimated to influence the expression of over 90% of human genes.
Alternative RNA splicing
The process of alternative splicing significantly enhances proteomic complexity, enabling cells to generate a vast array of proteins from a relatively modest number of genes. Beyond expanding the protein repertoire, alternative splicing plays a fundamental role in regulating gene expression, and it is tightly linked to various essential biological functions. Dysregulation of alternative splicing has been implicated in numerous diseases, including cancer, neurodegenerative disorders, and cardiovascular diseases, underscoring its importance in both normal physiology and disease pathology.
Our genomic analysis services can offer critical insights if you're looking for an in-depth analysis of alternative splicing events. By leveraging our expertise, we help you explore the complex landscape of alternative splicing and its impact on gene expression, providing a deeper understanding of molecular biology and disease mechanisms.
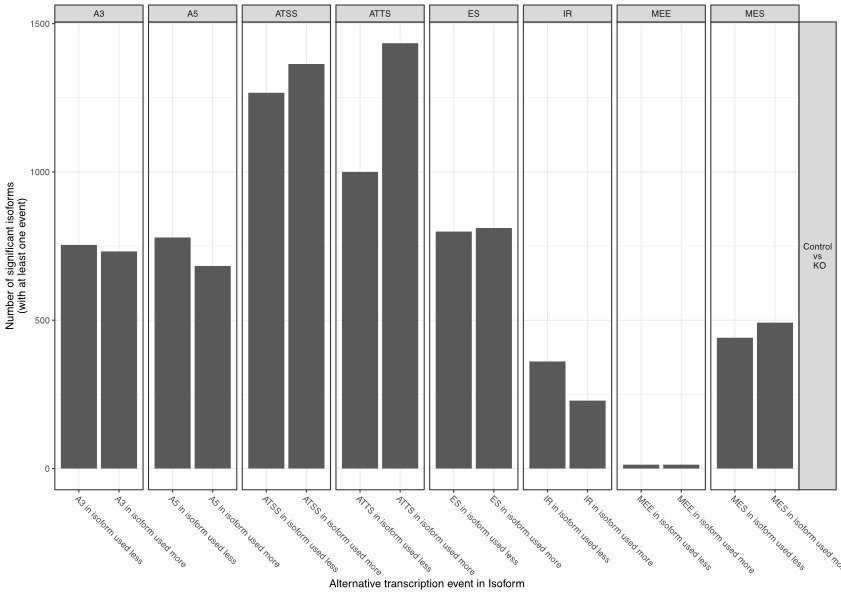
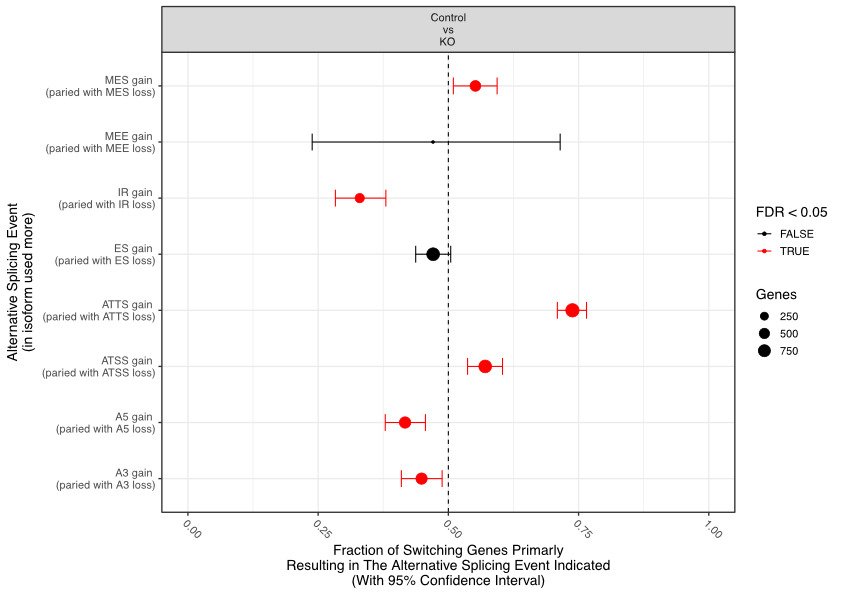
Types of Alternative Splicing
Alternative splicing is a highly versatile mechanism, resulting in several distinct forms that give rise to different mRNA and protein variants. Some of the most common types include:
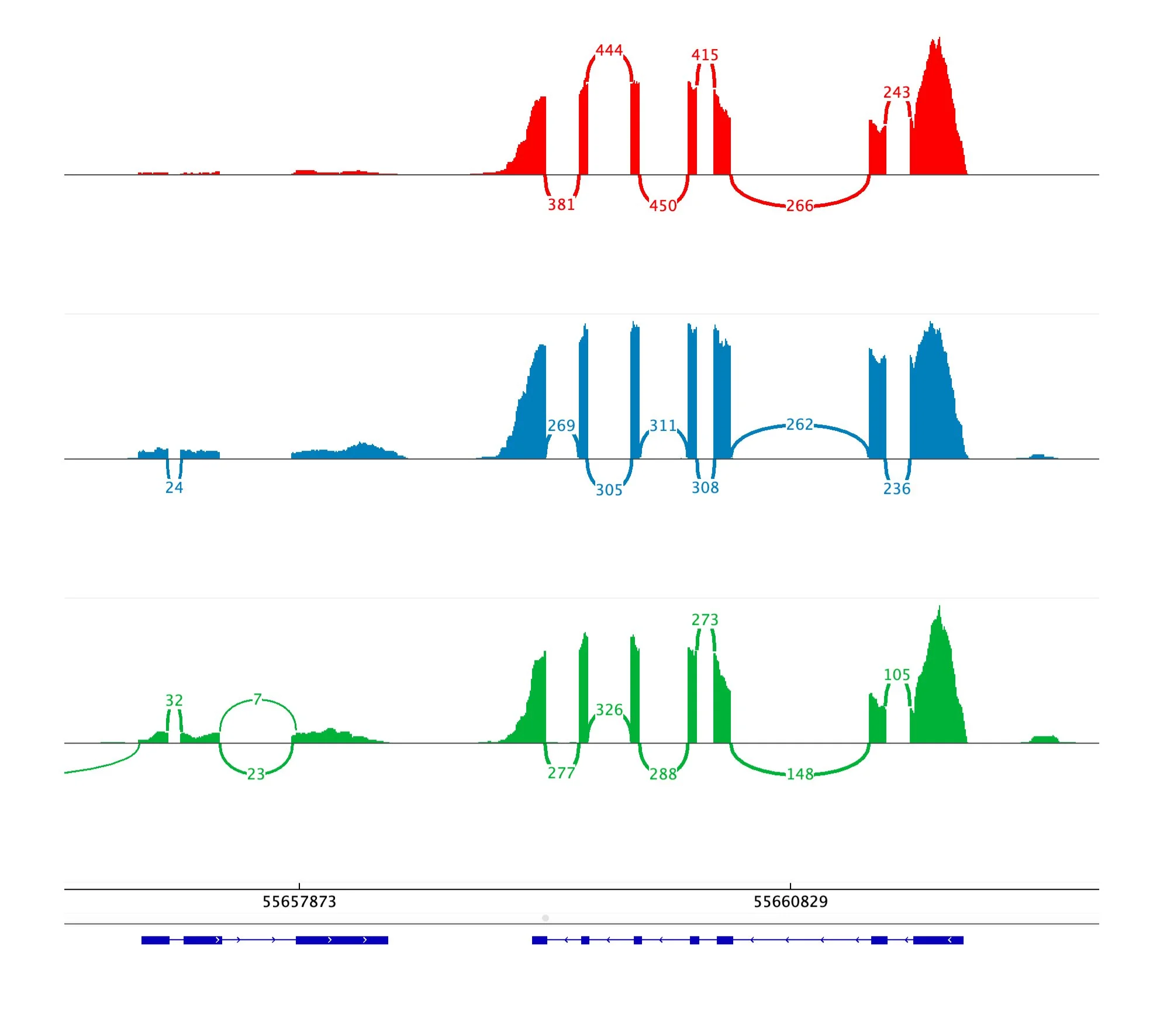
Exon Skipping: The most frequent form of alternative splicing, where specific exons are excluded from the final mature mRNA. This process alters the structure of the resulting protein by omitting key coding regions, which can significantly impact the protein's function, potentially introducing new or loss-of-function characteristics. Exon skipping is a critical mechanism in many biological processes and is often implicated in disease conditions when misregulated.
Intron Retention: Unlike typical splicing where introns are removed, this form leaves an intron intact within the mature mRNA. This inclusion of non-coding sequences can disrupt normal protein synthesis or lead to the production of a non-functional protein. Intron retention is more common in certain tissues and under specific developmental conditions and has been observed in cancers where it contributes to malignancy.
Alternative 5' or 3' Splice Sites: Splicing occurs at alternative donor (5') or acceptor (3') sites within the same exon. This variation leads to including shorter or longer regions of an exon in the mRNA, thereby producing proteins with altered N-terminal or C-terminal regions. This can subtly or dramatically affect the protein's stability, localisation, or interaction with other molecules, allowing cells to fine-tune protein function.
Mutually Exclusive Exons: In this type, only one of two possible exons is included in the final mRNA. Mutually exclusive exons provide a mechanism for generating proteins with different functional domains from the same gene. This is particularly important when alternative protein functions are needed in response to environmental cues or during various stages of development.
Each type of alternative splicing allows for a fine-tuned regulation of gene expression, contributing to the enormous diversity of proteins produced from a single gene. This variation in protein isoforms is essential for cellular differentiation, organismal development, and organisms' ability to adapt to environmental changes.
Please visit our blog for more detailed information about alternative splicing and its implications.